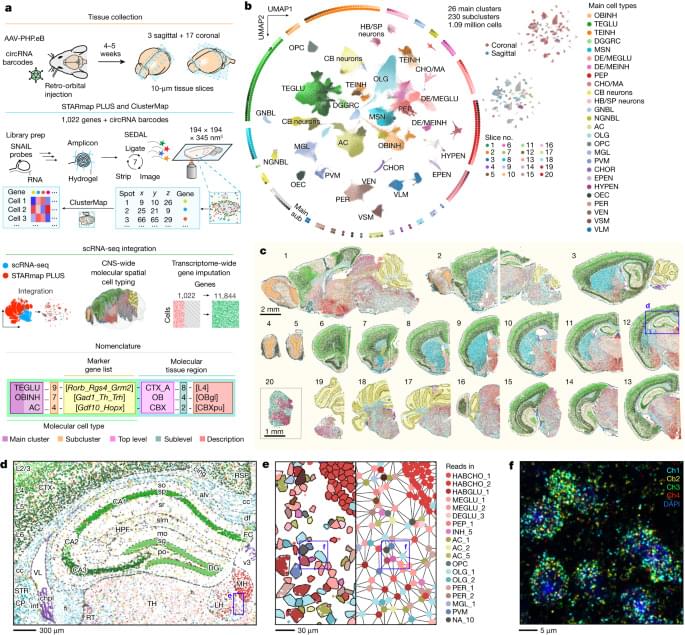
Stupendous paper on a new spatial transcriptomic atlas of mouse brain by Shi et al. from Xiao Wang’s group at MIT. They leverage their in situ sequencing method “STARmap PLUS” to profile 1,022 genes in 3D and map 1.09 million cells across the adult mouse brain and spinal cord. While they did not use the whole brain, instead opting for a series of thick sections at regular intervals, they still covered a lot of ground! Furthermore, they employed graph-theoretic computational methods to predict wider gene expression profiles of cells in their dataset, imputing single-cell expression profiles of 11,844 genes. This dataset/resource will serve the neurobiology community in elucidating the mechanistic workings of the brain!
In situ spatial transcriptomic analysis of more than 1 million cells are used to create a 200-nm-resolution spatial molecular atlas of the adult mouse central nervous system and identify previously unknown tissue architectures.