Apr 18, 2020
Genetic tracing ‘barcode’ is rapidly revealing COVID-19’s journey and evolution
Posted by Genevieve Klien in categories: biotech/medical, genetics
Drexel University researchers have reported a method to quickly identify and label mutated versions of the virus that causes COVID-19. Their preliminary analysis, using information from a global database of genetic information gleaned from coronavirus testing, suggests that there are at least six to 10 slightly different versions of the virus infecting people in America, some of which are either the same as, or have subsequently evolved from, strains directly from Asia, while others are the same as those found in Europe.
First developed as a way of parsing genetic samples to get a snapshot of the mix of bacteria, the genetic analysis tool teases out patterns from volumes of genetic information and can identify whether a virus has genetically changed. They can then use the pattern to categorize viruses with small genetic differences using tags called Informative Subtype Markers (ISM).
Applying the same method to process viral genetic data can quickly detect and categorize slight genetic variations in the SARS-CoV-2, the novel coronavirus that causes COVID-19, the group reported in a paper recently posted on the preliminary research archive, bioRxiv. The genetic analysis tool that generates these labels is publicly available for COVID-19 researchers on GitHub.
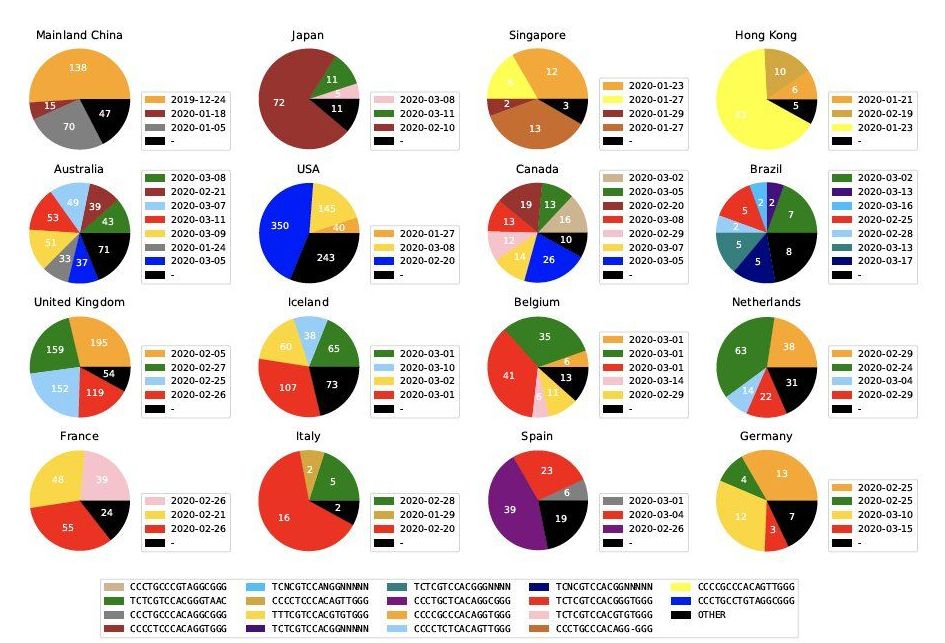


 Via Harvard David A. Sinclair “The coronavirus is part bat & part human virus. A new study says the Frankenstein event happened well before its transmission to humans. Wait, what? Humans first infected bats?
Via Harvard David A. Sinclair “The coronavirus is part bat & part human virus. A new study says the Frankenstein event happened well before its transmission to humans. Wait, what? Humans first infected bats?