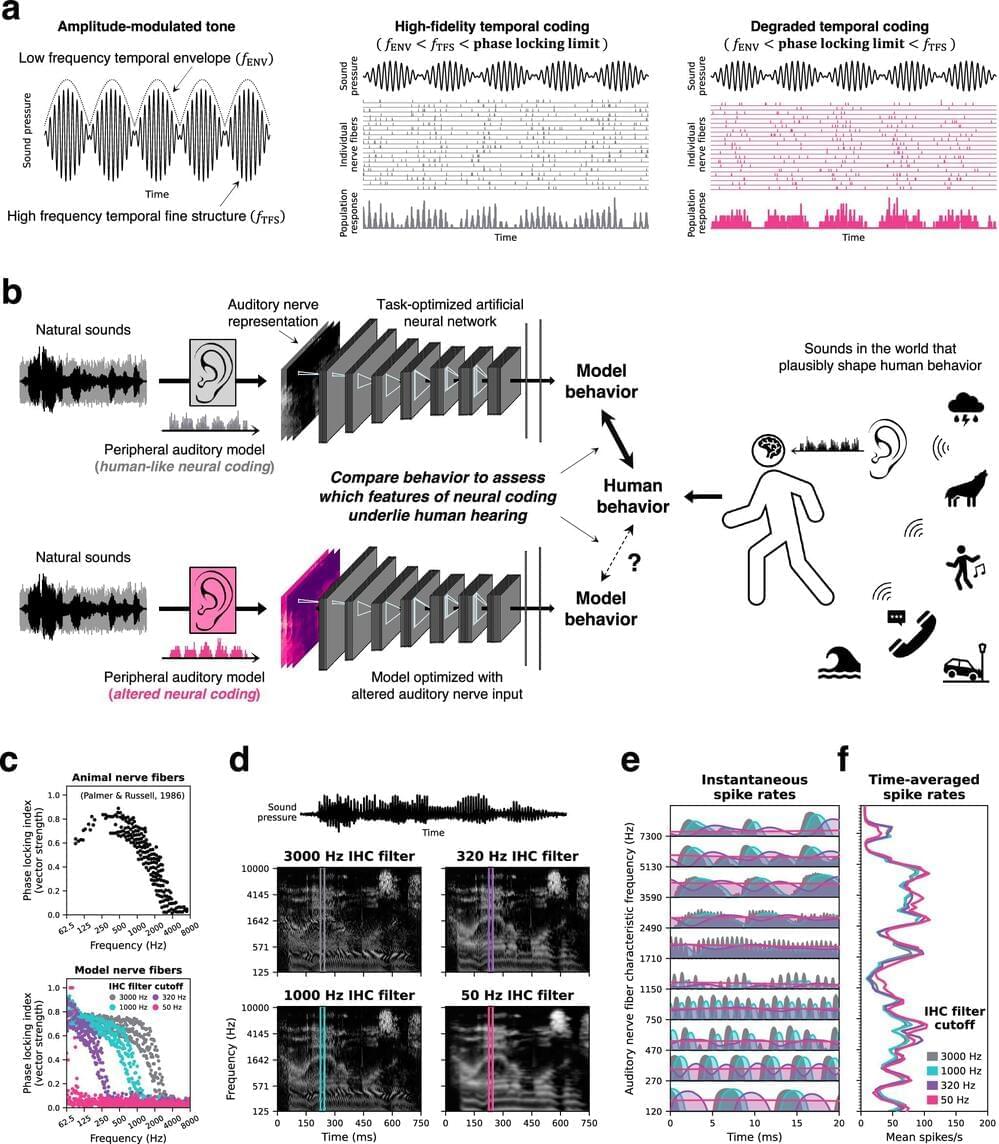
Back in the old days—the really old days—the task of designing materials was laborious. Investigators, over the course of 1,000-plus years, tried to make gold by combining things like lead, mercury, and sulfur, mixed in what they hoped would be just the right proportions. Even famous scientists like Tycho Brahe, Robert Boyle, and Isaac Newton tried their hands at the fruitless endeavor we call alchemy.
Materials science has, of course, come a long way. For the past 150 years, researchers have had the benefit of the periodic table of elements upon which to draw, which tells them that different elements have different properties, and one can’t magically transform into another. Moreover, in the past decade or so, machine learning tools have considerably boosted our capacity to determine the structure and physical properties of various molecules and substances.
New research by a group led by Ju Li—the Tokyo Electric Power Company Professor of Nuclear Engineering at MIT and professor of materials science and engineering—offers the promise of a major leap in capabilities that can facilitate materials design. The results of their investigation appear in Nature Computational Science.